How to guide: Filtering tool
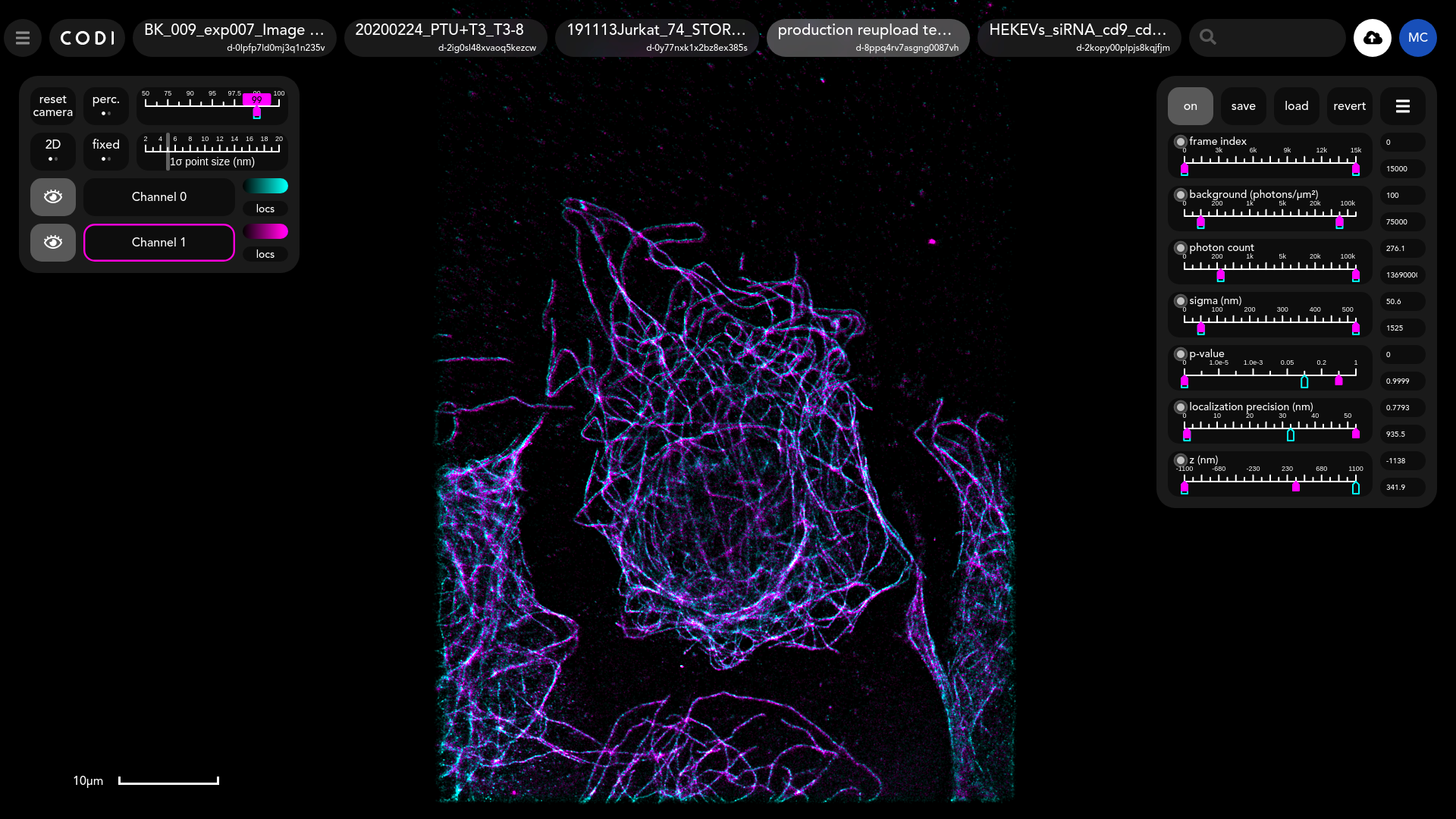
Tool view
Purpose of tool
Enable user to filter out points based on their properties and save the output for further analysis in workflow.
Contents
Input parameters
Multiple channels
Output results
Input parameters
The points in dSTORM acquisitions have the properties shown in the right-hand panel. Sliders can be used to filter out points by choosing ranges for these properties.
The checkbox in the top left corner of each slider is used to determine whether that property will be used to filter the points. The number of properties displayed depends on those present in the data.
The localisation algorithm runs a statistical fitting procedure on each localisation, producing a number of properties. Those available through the filtering tool are summarised here:
frame index: The frame in which the localisation was detected.
background [photons/μm^2]: density of photons from the background at the spot.
photon count: photons at the spot.
sigma [nm]: the standard deviation of the fitted point spread function at the spot. This filter acts at the same time on both the nimOS parameters Sigma X and Sigma Y.
p-value: smaller values correspond to a more reliable localisations. Normally used to filter out points with high p-value.
localisation precision [nm]: Estimation of the spatial uncertainty of the localisations. This filter acts at the same time on both the nimOS parameters Localisation precision X and Localisation precision Y, which are the Cramér–Rao lower bounds for the X and Y coordinates of the localisation.
z [nm] : The z-location of each spot. Only available for 3D datasets.
Multiple channels
If a dataset has multiple channels, then the set of channels to be filtered can be selected in the visualisation panel as shown below where two channels have been selected (highlighted borders). Individual channels can also be selected separately to apply different filters to each.
Output results
Once the filters have been selected, the current parameter settings can then be saved by clicking on the save button on the top right corner of the right hand side panel. These settings can be saved to the cloud (on CODI) or on the local machine. If saved on the local machine, a JSON file is downloaded showing the parameters that have been changed and the corresponding ranges. An example of a downloaded file is shown below. This same file can be loaded into the CODI filtering tool using the load button and that will automatically apply the filters to the image.
Related Articles
How to guide: Counting tool
Tool view Purpose of tool Counts the number of localisations or ‘bins’ by channel around a set of centre points - currently the centroids of a clustering result. Contents Inputs Results Outputs Inputs The centre points for counting come from a loaded ...How-to-guide: Temporal grouping tool
Tool view Purpose of tool In localisations-based super-resolution microscopy, each fluorescent molecule can emit ("blink") multiple times, depending on its properties and the super-resolution technique used. Those blinking events are localized ...How to guide: The CODI Desktop Uploader
Tool view Purpose of tool The CODI Desktop Uploader is a tool created to ease the transition between NimOS and CODI. The main objective of the application is to facilitate the uploading of NimOS generated datasets to the CODI cloud platform. It ...How to guide: Clustering tool
Tool view Purpose of tool The tool performs clustering on localisation data to find biological structures and quantify their statistical and morphological properties. The default clustering method is HDBSCAN. The cluster results can then be ...How to guide: Annotation tool (ROI)
Purpose of tool Select regions of interest (ROIs) in images. How to draw ROIs To begin drawing an ROI, click on one of the three green button depending on the shape you would like to draw. For the polygon: Click on the image, if possible around the ...